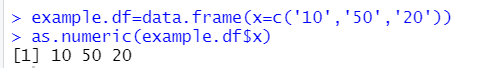R, the free and open source program for statistical computing, poses a substantial threat to the reproducibility of published research. This post explains the problem and introduces a solution.
The Problem: Packages
R itself has some reproducibility problems (see example in this footnote [1]), but the big problem is its packages: the addon scripts that users install to enable R to do things like run meta-analyses, scrape the web, cluster standard errors, format numbers, etc. The problem is that packages are constantly being updated, and sometimes those updates are not backwards compatible. This means that the R code that you write and run today may no longer work in the (near or far) future because one of the packages your code relies on has been updated. But worse, R packages depend on other packages. Your code could break after a package you don't know you are using updates a function you have never even used.
Example: dplyr
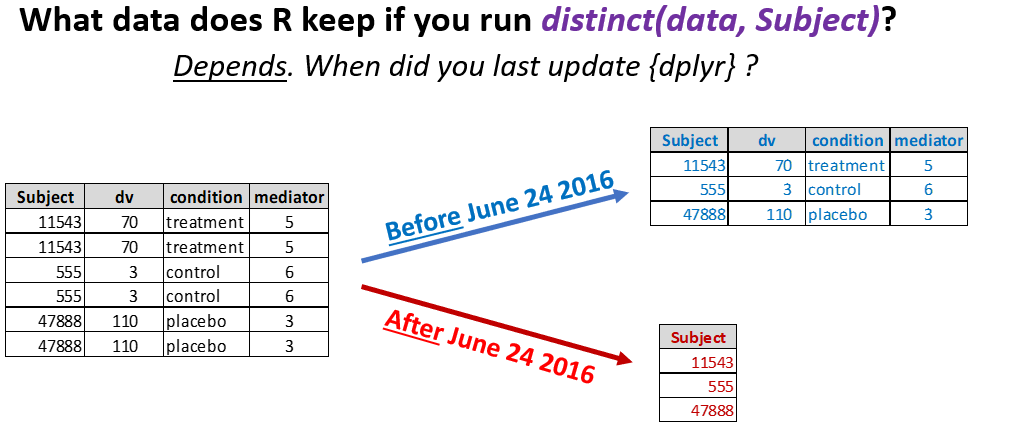
The very popular dplyr package is part of the tidyverse empire. It is used by hundreds of other packages, and by perhaps millions of scripts. The dplyr package is used to manipulate data. For example, its distinct() function eliminates rows with duplicate values in a dataset (e.g., with the same ID).
On June 24th, 2016, the creators of dplyr changed what this distinct() function does by default (I owe this example to Mark Brandt (.htm). Thanks Mark!). Before that date, distinct() would keep all variables in the dataset, but after that date it would only keep the variables you are checking to look for duplicates. The figure below illustrates:
The change probably broke every R script written before June 24, 2016 that relied on the function distinct(). And even scripts that do not use the distinct() function may stop working if they rely on a function from another package that uses distinct().
Packages get updated a lot
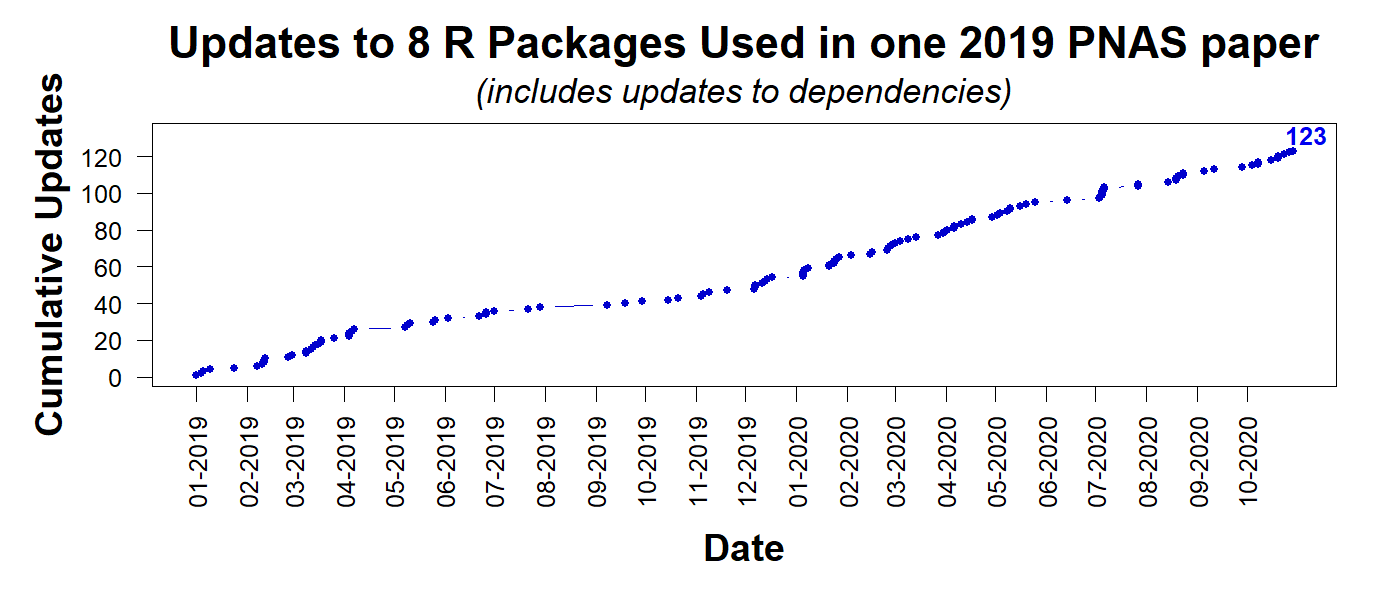
For illustration, consider the PNAS paper discussed in Colada[79]. The authors posted their R Code, allowing others to reproduce their results. At least that's the goal. The code relies on 8 packages. The figure below shows how many times those 8 packages, and their 'dependencies,' the packages those packages depend on, have been updated since January 1st 2019. As of Nov 17th 2020: 123 times.
We don't know the probability that a package update breaks a script; but, as a quick calibration, say it is 1%. The probability that the code for the PNAS paper breaks after 123 changes would be 71% [2]. If the base rate were just 1/1000 per change, then after 123 changes there is a ~12% chance of failure. How confident are you now that this code will run in 5 years?
Solution: groundhog
The solution to R's package problem is a new R package: groundhog.
If the package name seems strange, read this footnote [3].
All you need to do to fix this huge reproducibility problem is, instead of loading packages with the built-in library() command, load them with the groundhog.library() command.
That is it.
The groundhog.library() command takes two values. Like library(), you indicate which package you want to load. In addition, you enter a date. Any date. Groundhog will load the most recent version of that package, on CRAN, on that date. It will also load all dependencies of that package as current on that date.
So instead of this: library('dplyr')
You do this: groundhog.library('dplyr', '2016-06-20')
Now the distinct() function will do on every computer, and 'forever', the exact same thing [4].
Recommended usage
When starting a new script, choose a recent date (say first of the current or past month), and use it to load all packages. You can assign a variable to the date to make it easy to update. A sensible name is 'groundhog.day', but it can be anything you like.
So, my new scripts now start with something like this:
library(groundhog)
groundhog.day="2020-11-01"
pkgs=c('pwr','metafor','data.tables')
groundhog.library(pkgs, groundhog.day)
A nice feature of groundhog is that it makes 'retrofitting' existing code quite easy. If you come across a script that no longer works, you can change its library() statements for groundhog.library() ones, using as the groundhog.day the date the code was probably written (say when it was posted on the internet), and it may work again. For more details (.htm)
Bonus: no more install.packages()
When you use library() to ask for a package that you have not installed, you get an error. That's annoying.
When you use install.packages() to ask for a package you already have, the existing one gets deleted without warning. That's dangerous.
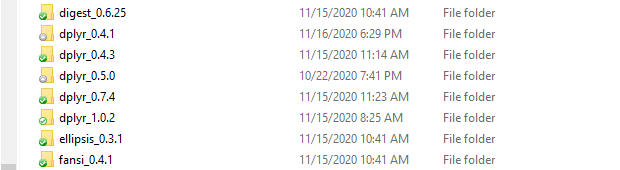
When you use groundhog.library() to ask for a package you don't have, it gets installed automatically, and saved alongside any existing versions of it. That's convenient.
You could have all 32 versions of dplyr side-by-side in your computer; groundhog will load the one you need for the date you enter. For example, this is what my groundhog folder looks like right now:
Other solutions
When I started working on groundhog I was aware of three existing solution to R's reproducibility problem. The packages renv (.htm), & checkpoint (.htm), and the more general solution: Docker (.htm). These solutions are sophisticated, powerful, and versatile, but they lack 3 features I thought wide adoption by researchers would require. An ideal solution would:
(1) Work within self-contained individual R scripts (e.g., not require projects, or additional files).
(2) Make it so that the code itself reveals which version of which packages were being used.
(3) Involve trivial adoption costs.
In this footnote, I discuss how these features are missing in existing solutions: [5].
Learn more
For most people: https://groundhogR.com
For those who self-identify as github users: https://github.com/CredibilityLab/groundhog
Credits
Groundhog was funded by the Wharton School of the University of Pennsylvania.
Like AsPredicted and ResearchBox, groundhog is brought to you by the Wharton Credibility Lab.
I wrote the first version of groundhog and then collaborated with Hugo Gruson, an evolutionary biologist (htm), to refine it, improve it, and turn it into a CRAN package.
To use groundhog:
install.packages('groundhog')
![]() Footnotes.
Footnotes.
- Regarding the irreproducibility of R coming from changes to R itself. Here is an example: With R 4.0.0, launched in 2020, there was a big change in a default of how data is read, breaking possibly millions of scripts. Here is a minimalistic example.
Let's have a dataset with a variable that is entered as text but you want to read as numeric (rather common situation):
example.df <- data.frame(x=c('10','50','20') )
as.numeric(example.df$x)If you run that in R-3.6.3 you get: 1,3,2 (!)

If you run that in R-4.0.0 you get a more sensible: 10,50,20
Same code, in the same program, R, gives you one result if you use the January 2020 version of R, and a different in the February 2020 version of R. The reason is that until R-4.0.0 string data was treated a 'factors', but now it is treated as character variables. [↩]
- The probability that it will stop working is 1-.99^123)=.7095 [↩]
- If you are too young to get the groundhog reference or otherwise have no memory of the 1990s, it's a 1993 movie in which Bill Murray's character experiences Groundhog Day over and over and over… .htm [↩]
- Well, nothing is forever, but it will work for long enough [↩]
-
renvworks at the R project rather than script level. Seerenv's "workflow" here: .htm. This means you need to create a new project for each self-contained R script you wish to write (something I don't currently do, and suspect many other researchers do not do either). Then, within a script in a project, code must be executed in a particular order for the installed packages to be saved to therenvcache library rather than the default library. The R code itself does not indicate which version of each package is being used. One must share a 'lockfile' alongside the .R scripts for the code to load the right packages in someone else's computer.checkpoint also requires you work in projects (.Rproj) rather than individual stand-alone .R scripts. See their instructions for details . I believe
checkpointhas some potential reproducibility shortcomings by relying on MRAN, Microsoft's archive copy of CRAN, to determine which version of a package to install. For a given date, CRAN holds multiple versions of the same package, and the one that is served depends on the operating system and R version making the request, so, the same code relying on checkpoint could load one package on R-3.5.1 and a different one on R-4.0.3, one package on UNIX and a different on Windows, etc.groundhogalso relies on MRAN, but it decides which version of the package is to be loaded independently. For any R version, and any operating system the same package version is loaded for a given date. I believe checkpoint will also need to re-install every package you use for in a new project, even if you have installed it before, but am not sure.Docker works by recreating an entire computing environment (including operating system). It's a great solution for many big problems, but an overkill -in my opinion- to simply ensure that a few lines of R code are reproducible. Instead of sharing just my R scripts (almost always <1000 lines of code), I need to share also an image file that includes all packages, R, the data used, R-Studio, and the operating system. I would personally recommend that very large efforts, for example, a Many Labs project, be shared in a Docker, but also as individual files relying on groundhog. Indeed, Docker and groundhog can be used as complements rather than substitutes.
Specifically, I am looking into creating virtual machines that come with all R Versions and all versions of all packages installed, providing stronger longer term reproducibility. This would allow a user to download a single virtual machine, and use groundhog() to load any package for any R Version without needing to install anything else. If you are interested in collaborating in creating this solution, please get in touch. I suspect Docker is not necessary for this, and VirtualBox or VMWare would provide an easier solution [↩]