This post shows how to run simulations (loops) in R that can go 50 times faster than the default approach of running code like: for (k in 1:100) on your laptop. Obviously, a bit of a niche post.
There are two steps.
Step 1 involves running parallel rather than sequential loops [1].
This step can increase your speed about 8 fold.
Step 2 involves using a virtual computer instead of your lame laptop.
This step can increase speed about another 7 fold.
Both steps combined, then, lead to about 50 fold speed increase [2].
If you already know how to run parallel loops on a virtual desktop, check out step 2 anyway, it suggests a faster, simpler, cheaper, more flexible alternative to Amazon's AWS.
A motivating example: simulation to evaluate whether likert scales can be t-tested
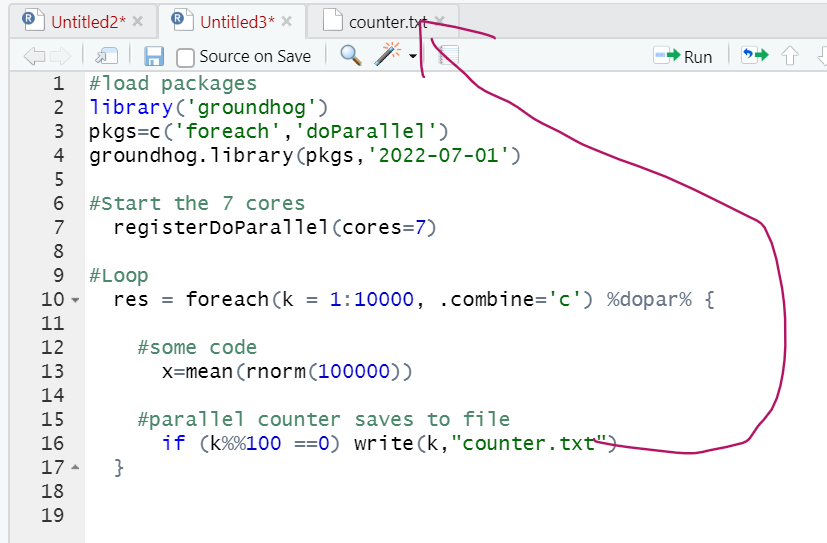
To have a concrete loop to contrast sequential vs parallel, let’s consider running simulations to see if it is OK to run a t-test on a likert scale. Specifically, the simulation generates two random samples with possible Likert values (1, 2, 3, 4 or 5), runs a t-test on these two samples, and saves the p-value. The loops repeat this simulation 8000 times, saving the p-value each time. Once all the simulations are completed, we can check how many of these p-values are smaller than .05 (should be 5%). This example is good in that it is simple and at least somewhat interesting, but the example is bad in that the simulation is not very demanding so it runs fast enough without bothering with parallel processing. This code runs in seconds. But more demanding simulation can take hours to run. In any case, the simulation would be like this:
set.seed(111)
p=c()
for (k in 1:8000) {
x1=sample(1:5 , size=30,replace=TRUE)
x2=sample(1:5 , size=30,replace=TRUE)
p[k]=t.test(x1,x2)$p.value
}
The loop produces a vector with 8000 p-values. The false-positive rate is the % of those 8000 that are p<.05. We can compute it with sum(p<.05)/8000. We obtain 4.94%. Close enough to 5%. So, turns out t-testing likert scales is fine [3].
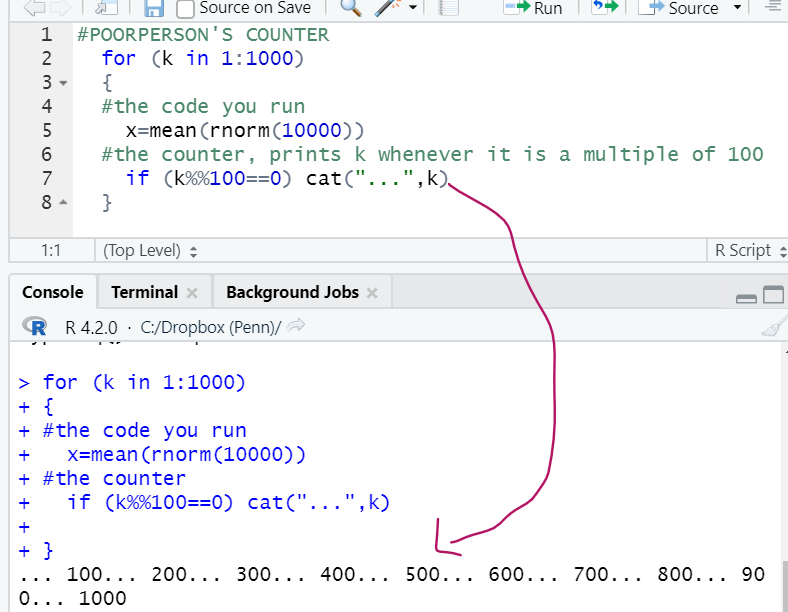
for() is kind of a dumb loop.
Dumb in that while our computers have the capacity to do many things at the same time, we are asking it to do only one thing, one loop, at a time. Computers come with multiple core processors. To know how many your computer has you can run parallel::detectCores()
My computer has 8. But I only used 1 to learn about likert scales above.
One of my 8 processors run 8000 simulations, the other 7 did nothing.
Imagine you visit a French restaurant with your favorite cousin. The waiter takes forever to come take your order, you are getting upset till you realize there is only one waiter working. But then, you realize there are 7 other waiters, all on break. You become upset again, and quickly come up with an idea that would speed things up.

Fig 1. Visual approximation of laptop while running a for() loop
That’s right. Have all the waiters instead of only one of them do some work.
Same with R, let’s have each of the 8 cores do 1000 simulations, at the same time, instead of one core do 8000 one after the other.
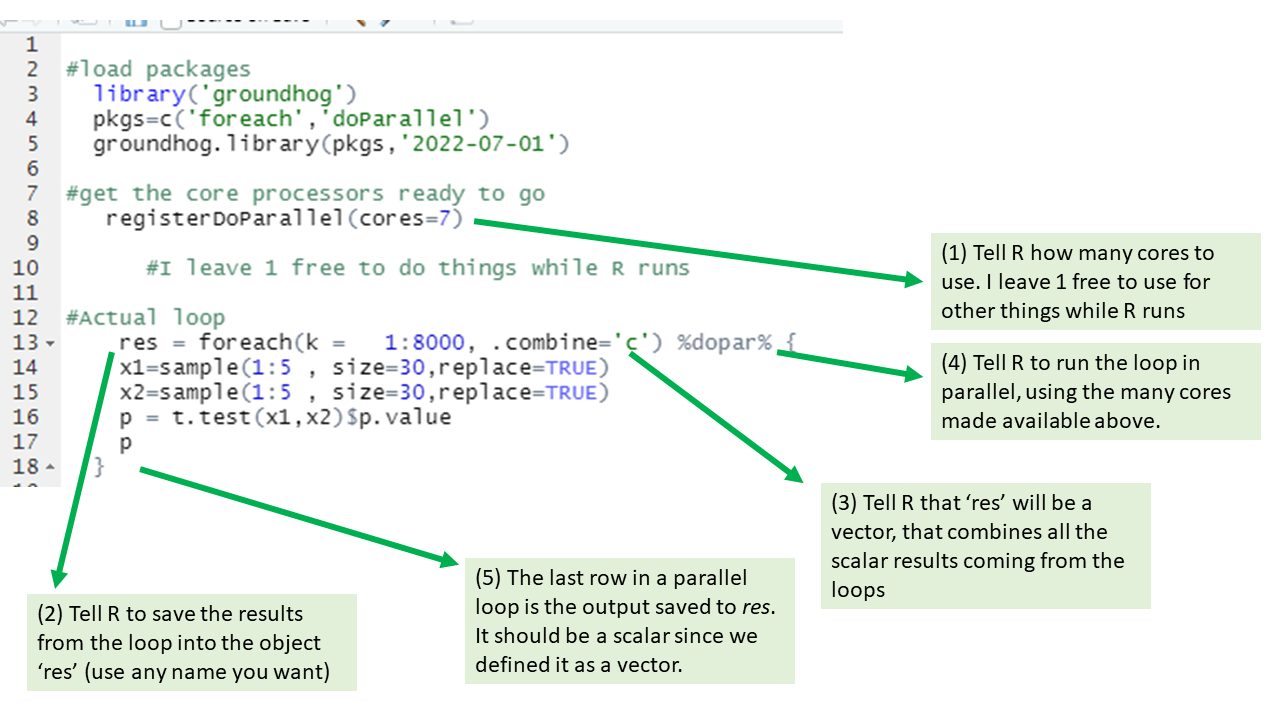
Step 1. Running parallel loops in R
The figure below has annotated code for running the same loop we saw above, but in parallel.
This loop will run about 8 times faster than the for() loop, minus the fixed time cost of setting up all the parallel processing. For fast tasks, parallel processing is not worth it.
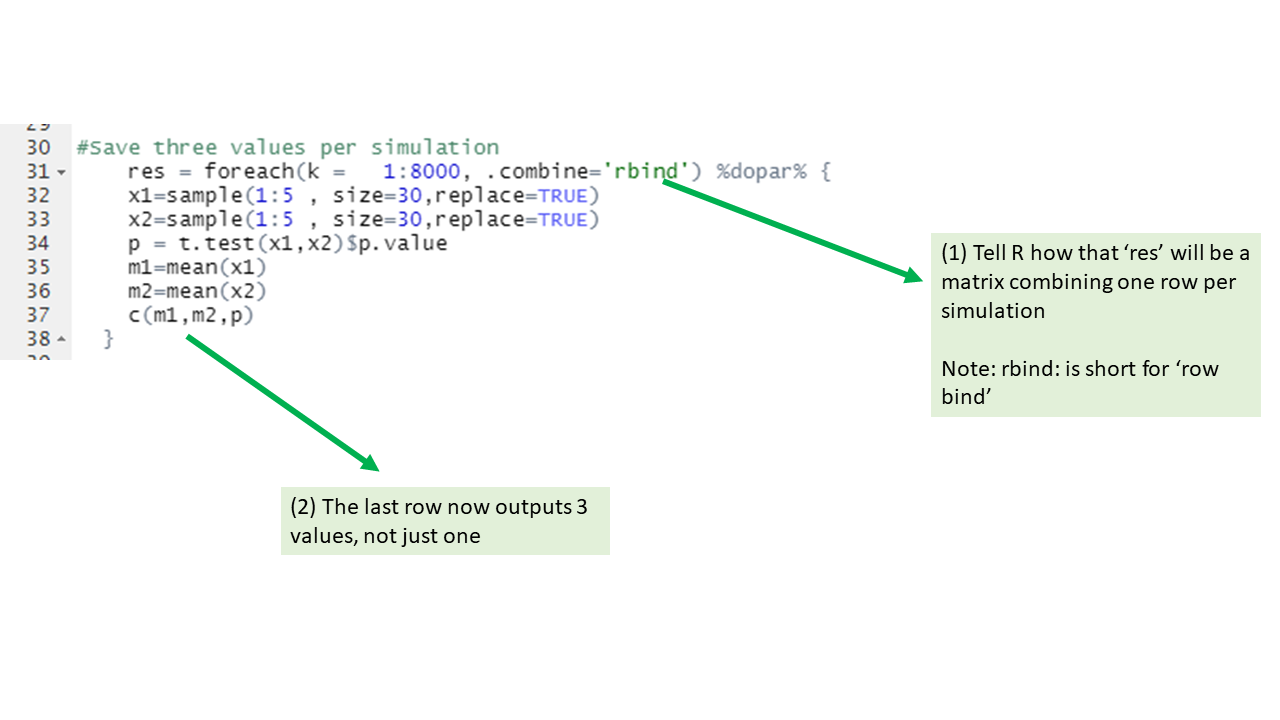
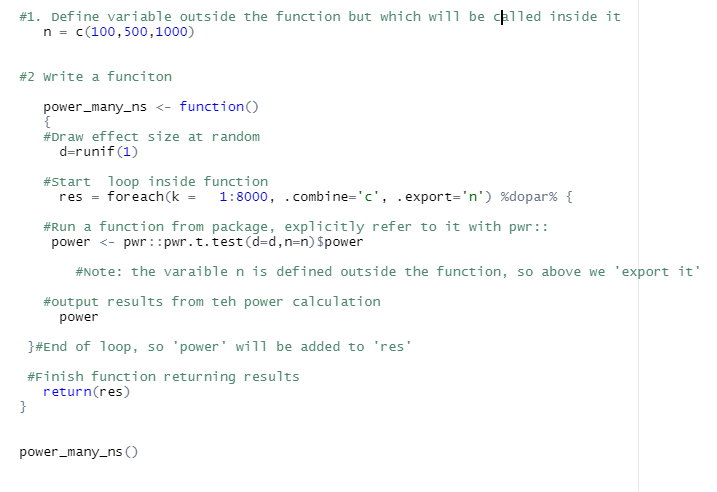
Intermediate level tips
If you start using parallel loops, these extra pieces of info may be useful
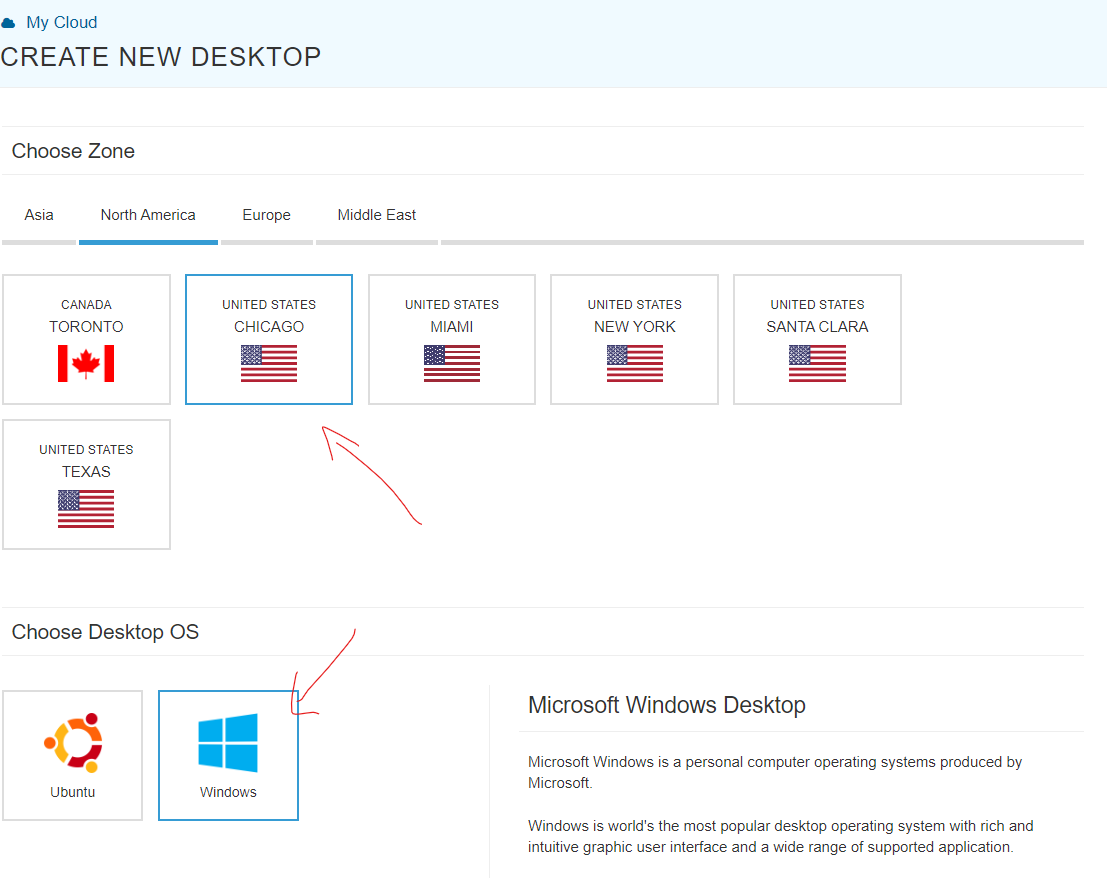
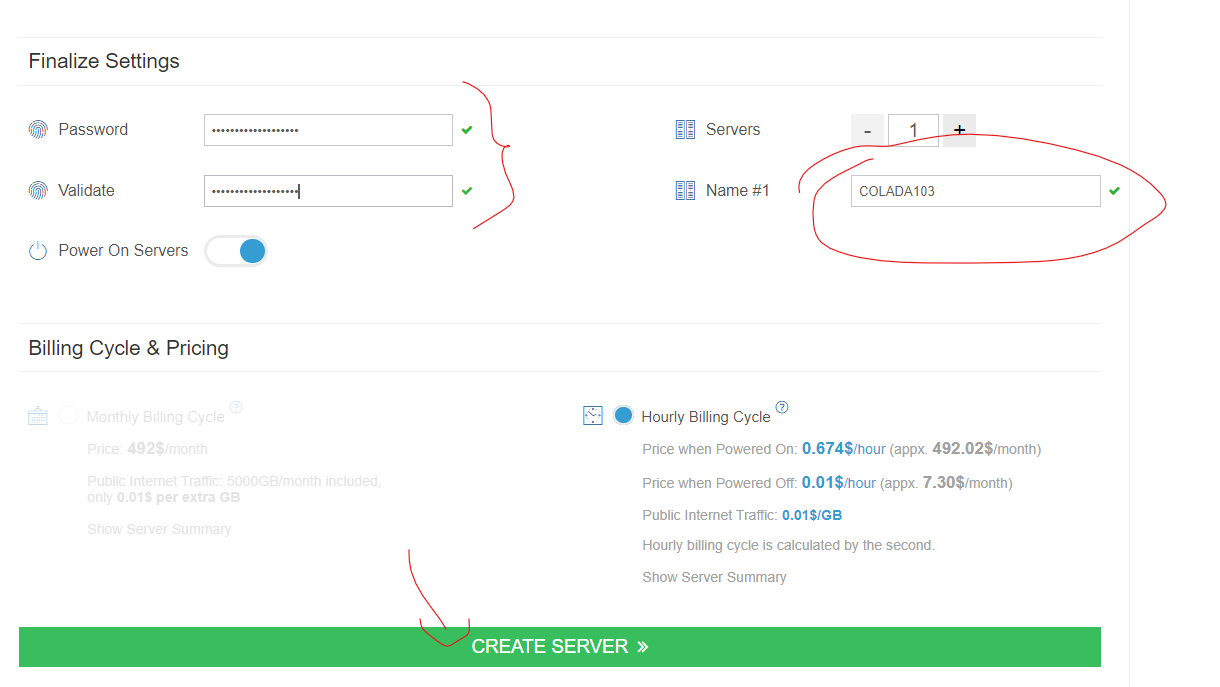
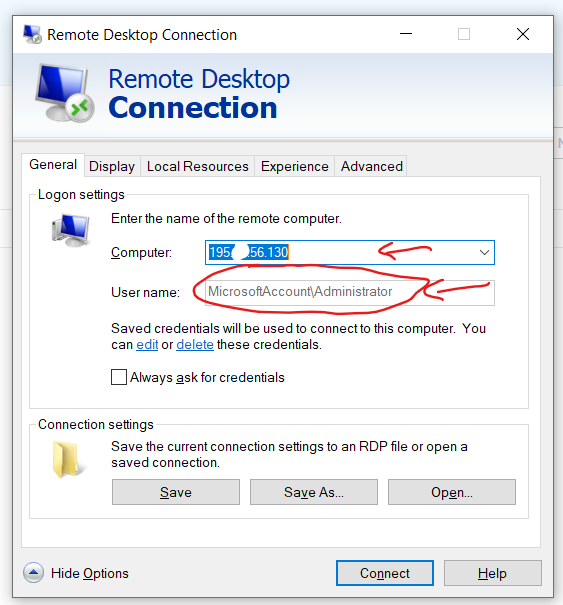
Step 2. Running parallel loops on a virtual desktop If you haven't learned how to use AWS, do not bother. To use it, go to http://kamatera.com and then: 1. Create an account 2. My cloud→Servers→Create New Desktop 3. Choose any location and presumably Windows machine. 4. Select the specs for your virtual computer I am choosing 48 cores here, the key component, and some RAM and HD space which are usually not too relevant for loops. Kamatera sets a limit to the specs based on how much it would cost to leave the virtual machine on for a full month. That limit is about $350. if you don't modify that cap (which you can do by emailing support) you will have to choose fewer than 50 cores. [5]. 5 Choose a name and password for the virtual computer. Note: Make sure to choose ‘hourly billing’ so you don’t get charged when you don’t use it It takes about 5 literal minutes for the virtual computer to be created. So don't worry if it does not immediately appear. 6. Then start the virtual computer clicking on “Action->power on” 7. Connect with remote desktop app in your computer, entering the IP address shown in the Kamatera site Look at the arrow to see what to use as user name Whem prompted for the password, enter the one you just set above. Mac user? connect to the windows machine from your Mac with: 8. Now you have a fully functional virtual windows machine. Once you are connected this is your virtual machine, you can do everything in it you do on your pc. To install R Studio, for example, just open the browser and download it. You can also copy-paste code from your computer to the virtual one and vice versa. 9. Bonus: github
Your laptop probably has at most 8 cores, limiting the benefits of parallel loops. If you run them on a virtual computer on the cloud with more cores, things will naturally go even faster. I will show you how to setup a virtual desktop with ~50 cores. You have to pay, but it is cheap, <$1 (one dollar) per hour of computations. I first used Amazon's AWS to run R on the cloud, but I recently came across a way better alternative kamatera.com.
If you have learned it, you probably will prefer Kamatera, it is much easier to use, way faster to set up, and flexible (e.g., you can easily update R and R Studio in it).
(sorry, life is hard sometimes)
You probably want type=’burstable’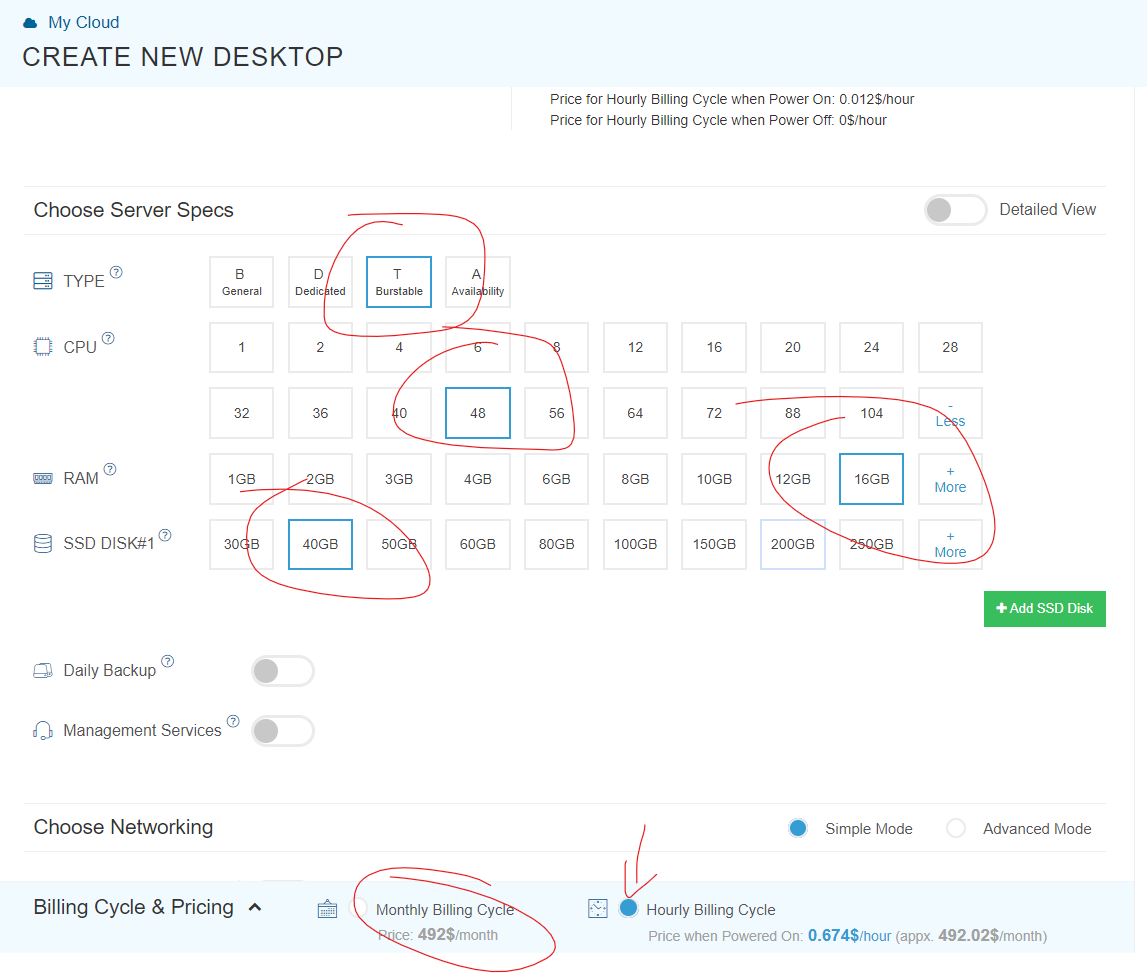
Kamatera actually emails you once they have it ready.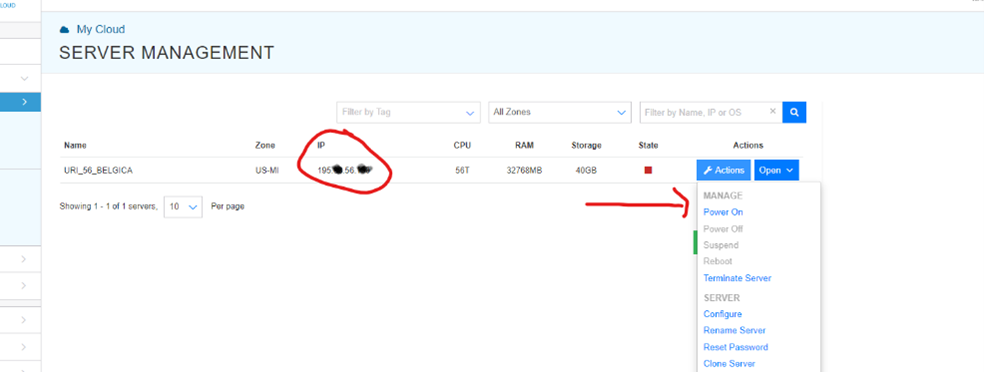
https://apps.apple.com/us/app/microsoft-remote-desktop/id1295203466?mt=12
If you use github, that's by far the easiest way to send files back and forth. Install it in the virtual machine, and just pull before you work and push after you do. Code and results will be synced.![]()
Footnotes.